Pocket K No. 54: Plant Breeding Innovation: CRISPR-Cas9
The CRISPR-Cas9 system is a plant breeding innovation that uses site-directed nucleases to target and modify DNA with great accuracy.1,2 Developed in 2012 by scientists from the University of California, Berkeley, CRISPR-Cas9 has received a lot of attention in recent years due to its range of applications, including biological research, breeding and development of agricultural crops and animals, and human health applications.1,2 These include gene silencing, DNA-free CRISPR-Cas9 gene editing, homology-directed repair (HDR), and transient gene silencing or transcriptional repression (CRISPRi).1,3,4,5
What is CRISPR and how it works
CRISPR, or Clustered Regularly Interspaced Short Palindromic Repeats, is an integral part of a bacterial defense system. It is also the basis of the CRISPR-Cas9 system.1,2,3,5
The CRISPR molecule is made up of short palindromic DNA sequences that are repeated along the molecule and are regularly-spaced. Between these sequences are “spacers”, foreign DNA sequences from organisms that have previously attacked the bacteria. The CRISPR molecule also includes CRISPR-associated genes, or Cas genes. These encode proteins that unwind DNA, and cut DNA, called helicases and nucleases, respectively.1,6
The CRISPR immune system protects the bacteria from repeated virus attacks thru three steps:
1. Adaptation – When DNA from a virus invades the bacteria, the viral DNA is processed into short segments and is made into a new spacer between the repeats. These will serve as genetic memory of previous infections.
2. Production of CRISPR RNA – The CRISPR sequence undergoes transcription, including spacers and Cas genes, creating a single-stranded RNA. The resulting single-stranded RNA is called CRISPR RNA, which contains copies of the invading viral DNA sequence in its spacers.
3. Targeting – The CRISPR RNAs will identify viral DNA and guide the CRISPR-associated proteins to them. The protein then cleaves and destroys the targeted viral material.
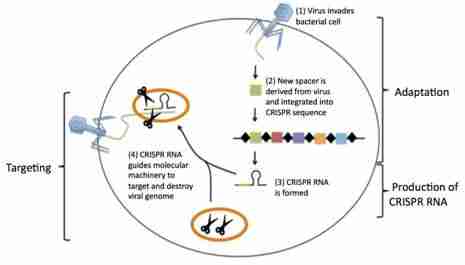
Figure 1. The steps of CRISPR-mediated immunity6
Scientists make use of the CRISPR-Cas9 systems’ recognition of specific DNA sequences and apply it in the process of development of improved crops. Instead of viral DNA as spacers, scientists design their own sequences, based on their specific gene of interest. If a gene’s sequence known, it can be easily used in CRISPR. It will then act just like a spacer for the system and guide the Cas9 protein to a DNA matching sequence.1,6
Figure 2. Mechanism of Gene Editing of CRISPR/Cas96
CRISPR-Cas9 allows researchers to perform the following:
Gene Knock-Out
Gene silencing using CRISPR starts with the use of a single guide RNA (sgRNA) to target genes and initiate a double stranded break using the Cas9 endonuclease. These breaks are then repaired by an innate DNA repair mechanisms, the non-homologous end-joining (NHEJ). However, NHEJ is error-prone and results in genomic deletions or insertions, which then translates into permanent silencing of the target gene. 4,7,8
DNA-Free Gene Editing
CRISPR can be used for DNA-free gene editing without the use of DNA vectors, requiring only RNA or protein components. A DNA-free gene editing system can be a good choice to avoid the possibility of undesirable genetic alterations due to the plasmid DNA integrating at the cut site or random vector integrations.4,7
Gene Insertions or “Knock-ins”
The CRISPR-induced double-strand break can also be used to create a gene “knock-ins” by exploiting the cells’ homology-directed repair. The precise insertion of a donor template can alter the coding region of a gene. Previous studies have demonstrated that single-stranded DNA can be used to create precise insertions using CRISPR-Cas9 system.4,7,8
Transient Gene Silencing
By modifying the Cas9 protein so it cannot cut DNA, transient gene silencing or transcriptional repression can also be done. The modified Cas9, led by a guide RNA, targets the promoter region of a gene and reduces transcriptional activity and gene expression. Transient activation or upregulation of specific genes can be effectively done.4,7
CRISPR - Cas9 Applications
Researchers have found that the CRISPR - Cas9 system can be applied to nearly every organism. Early studies using CRISPR - Cas9 for gene editing have focused on crops important for agriculture. It was realized early on that the system could be used in crops to improve traits, such as yield, plant architecture, plant aesthetics, and disease tolerance.
dense and erect panicle1 (DEP1) gene in the Indica rice line IR58025B. Improvements
in yield-related traits, such as dense and erect panicles and reduced plant height,were observed in the mutant plants produced.9
A team of researchers from the Chinese Academy of Agricultural Sciences led by Yupeng Cai also used the CRISPR-Cas9 system to induce mutations on GmFT2a, an integrator in the photoperiod flowering pathway of soybean. The developed soybean plants showed late flowering, resulting in increased vegetative size. The mutation was also found to be stably inherited in the following generation.10
Researchers from Beijing Key Laboratory of Vegetable Germplasm Improvement, led by Shouwei Tian used CRISPR-Cas9 to target ClPDS, the phytoene desaturase in watermelon, to achieve the albino phenotype. All genome-edited watermelons harbored mutations in ClPDS and showed full or mosaic albino phenotype. This study served as a proof of concept of using the CRISPR-Cas9 system in watermelon breeding.11
Xanthomonas citri subsp. citri (Xcc), a serious disease of citrus, through CRISPR-Cas9.
The team targeted the promoter of the CsLOB1 gene, which promotes canker development in citrus. The developed lines showed enhanced resistance to citrus canker compared to wild types.12
Cold Spring Harbor Laboratory, together with various research institutions, also used CRISPR-Cas9 to generate mutations in the flowering suppressor SELF-PRUNING5G (SP5G) in tomato to manipulate photoperiod response. The mutations brought about by CRISPR-Cas9 caused rapid flowering and enhanced the compact growth habit of field tomatoes, resulting in a quick burst of flower production and early yield.13
CRISPR-Cas9 has also allowed generation of animals suitable for human disease modeling. The team of Yuyu Niu from Yunnan Key Laboratory of Primate Biomedical Research applied CRISPR-Cas9 via coinjection of Cas9 mRNA and sgRNAs into one-cell-stage embryos. The team generated CRISPR-edited cynomolgus monkeys for brain disorders that cannot be fully studied in mice.14
15
US scientists are also studying the use of CRISPR for treating the Human Immunodeficiency Virus (HIV). They used CRISPR to edit the HIV genome out of immune cells, called T cells, from an HIV patient. Scientists found that CRISPR can prompt the HIV virus to mutate. However, more studies are still needed before CRISPR can be used to treat HIV.6
CRISPR has played a huge part in the increase in genome editing studies in recent years. The system has broad applications in plant and animal improvement, as well as in the medical field. As a relatively young technique, various discoveries and innovations for its efficient use in wider applications are in the offing.
References
- Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D.A., and Horvath, P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 (5819): 1709–1712.
- Horizon Discovery. 2016. CRISPR/CRISPR Cas9. https://www.horizondiscovery.com/gene- editing/crispr.
- Bolotin, A., Quinquis, B., Sorokin, A.,and Ehrlich, S.D. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151 (8): 2551–2561.
- Dharmacon. 2016. Gene Editing. http://dharmacon.gelifesciences.com/applications/gene-editing/.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., and Charpentier, E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096): 816–821.
- Harvard University. 2015. CRISPR: A game-changing genetic engineering technique. http://sitn.hms.harvard.edu/flash/2014/crispr-a-game-changing-genetic-engineering-technique/.
- Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121): 819–823.
- AddGene. 2014. CRISPR History and Background. https://www.addgene.org/crispr/reference/history/.
- Wang, Y., Geng, L., Yuan, M., Wei, J., Jin C., Li, M., Yu K., Zhang Y., Jin, H., Wang, E., Chai, Z., Fu, X., Li, X. 2017. Deletion of a target gene in Indica rice via CRISPR/Cas9. Plant Cell Reports 36 (8): 1333–1343.
- Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., Jiang, B., Han, T. and Hou, W. 2017. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnology Journal. http://onlinelibrary.wiley.com/doi/10.1111/pbi.12758/full.
- Tian, S., Jiang, L., Gao, Q., Zhang, J., Zong, M., Zhang, H., Ren, Y., Guo, S., Gong, G., Liu, F., Xu, Y. 2016. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Reports 36 (3): 399–406.
- Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., Yao, L. and Zou, X. 2017. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnology Journal. http://onlinelibrary.wiley.com/doi/10.1111/pbi.12733/full.
- Soyk, S., Müller, N.A., Park, S.J., Schmalenbach, I., Jiang, K., Hayama, R., Zhang, L., Van Eck, J., Jiménez-Gómez, J.M., and Lippman, Z.B. 2017. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nature Genetics, 49 (1): 162–168.
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., Xiang AP., Zhou J., Guo X., Bi Y., Si C., Hu B., Dong G., Wang H., Zhou Z., Li T., Tan T., Pu X., Wang F., Ji S., Zhou Q., Huang X., Ji W., Sha J. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156 (4): 836-843.
- Xie F., Ye L., Chang J.C., Beyer A.I., Wang J., Muench M.O. and Kan Y.W. 2014. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Research 24 (9): 1526–1533.