Abstract
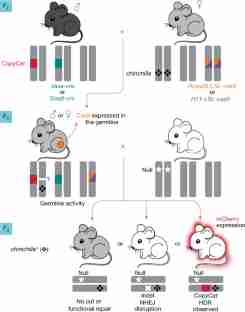
A gene drive biases the transmission of one of the two copies of a gene such that it is inherited more frequently than by random segregation. Highly efficient gene drive systems have recently been developed in insects, which leverage the sequence-targeted DNA cleavage activity of CRISPR–Cas9 and endogenous homology-directed repair mechanisms to convert heterozygous genotypes to homozygosity1,2,3,4. If implemented in laboratory rodents, similar systems would enable the rapid assembly of currently impractical genotypes that involve multiple homozygous genes (for example, to model multigenic human diseases). To our knowledge, however, such a system has not yet been demonstrated in mammals. Here we use an active genetic element that encodes a guide RNA, which is embedded in the mouse tyrosinase (Tyr) gene, to evaluate whether targeted gene conversion can occur when CRISPR–Cas9 is active in the early embryo or in the developing germline. Although Cas9 efficiently induces double-stranded DNA breaks in the early embryo and male germline, these breaks are not corrected by homology-directed repair. By contrast, Cas9 expression limited to the female germline induces double-stranded breaks that are corrected by homology-directed repair, which copies the active genetic element from the donor to the receiver chromosome and increases its rate of inheritance in the next generation. These results demonstrate the feasibility of CRISPR–Cas9-mediated systems that bias inheritance of desired alleles in mice and that have the potential to transform the use of rodent models in basic and biomedical research.
Access options
Subscription info for Chinese customers
We have a dedicated website for our Chinese customers. Please go to naturechina.com to subscribe to this journal.
Rent or Buy article
Get time limited or full article access on ReadCube.
from$8.99
All prices are NET prices.
Data availability
All genotyping data for F3 offspring of constitutive crosses and F4 offspring of germline conditional crosses is available at Zenodo with the identifier https://doi.org/10.5281/zenodo.2003087. Annotated sequence data for the TyrCopyCat transgene is available in GenBank with the accession number MK160997.
Change history
08 January 2020
An Amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
-
1.
Gantz, V. M. & Bier, E. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015).
-
2.
Gantz, V. M. et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743 (2015).
-
3.
Hammond, A. et al. A CRISPR–Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016).
-
4.
Kyrou, K. et al. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018).
-
5.
Gantz, V. M. & Bier, E. The dawn of active genetics. BioEssays 38, 50–63 (2016).
-
6.
Gould, F. Broadening the application of evolutionarily based genetic pest management. Evolution 62, 500–510 (2008).
-
7.
Esvelt, K. M., Smidler, A. L., Catteruccia, F. & Church, G. M. Emerging technology: concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 (2014).
-
8.
Mao, Z., Bozzella, M., Seluanov, A. & Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 7, 1765–1771 (2008).
-
9.
Miyaoka, Y. et al. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 6, 23549 (2016).
-
10.
Xu, X.-R. S., Gantz, V. M., Siomava, N. & Bier, E. CRISPR/Cas9 and active genetics-based trans-species replacement of the endogenous Drosophila kni-L2 CRM reveals unexpected complexity. eLife 6, e30281 (2017).
-
11.
Yokoyama, T. et al. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 18, 7293–7298 (1990).
-
12.
Yen, S.-T. et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol. 393, 3–9 (2014).
-
13.
Miyagishi, M. & Taira, K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20, 497–500 (2002).
-
14.
Boshart, M. et al. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41, 521–530 (1985).
-
15.
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
-
16.
Chiou, S.-H. et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 29, 1576–1585 (2015).
-
17.
Beermann, F. et al. Rescue of the albino phenotype by introduction of a functional tyrosinase gene into mice. EMBO J. 9, 2819–2826 (1990).
-
18.
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997).
-
19.
Goedecke, W., Eijpe, M., Offenberg, H. H., van Aalderen, M. & Heyting, C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat. Genet. 23, 194–198 (1999).
-
20.
Gallardo, T., Shirley, L., John, G. B. & Castrillon, D. H. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-cre. Genesis 45, 413–417 (2007).
-
21.
Sadate-Ngatchou, P. I., Payne, C. J., Dearth, A. T. & Braun, R. E. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738–742 (2008).
-
22.
de Rooij, D. G. & Grootegoed, J. A. Spermatogonial stem cells. Curr. Opin. Cell Biol. 10, 694–701 (1998).
-
23.
Pepling, M. E. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 44, 622–632 (2006).
-
24.
Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R. Soc. Lond. B 270, 921–928 (2003).
-
25.
Deredec, A., Burt, A. & Godfray, H. C. J. The population genetics of using homing endonuclease genes in vector and pest management. Genetics 179, 2013–2026 (2008).
-
26.
Unckless, R. L., Clark, A. G. & Messer, P. W. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205, 827–841 (2017).
-
27.
Noble, C., Olejarz, J., Esvelt, K. M., Church, G. M. & Nowak, M. A. Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 (2017).
-
28.
Marshall, J. M., Buchman, A., Sánchez C, H. M. & Akbari, O. S. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci. Rep. 7, 3776 (2017).
-
29.
Noble, C., Adlam, B., Church, G. M., Esvelt, K. M. & Nowak, M. A. Current CRISPR gene drive systems are likely to be highly invasive in wild populations. eLife 7, e33423 (2018).
-
30.
Jensen-Seaman, M. I. et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14, 528–538 (2004).
Acknowledgements
We thank K. Hanley for the DNA extraction protocol; A. Green and A.-C. Chen for genotyping assistance; M. Tran for laser-capture microdissection in an effort to genotype spermatogonia; P. Jain for assistance with fibroblast transfection; H. Cook-Andersen and M. Wilkinson for conversations about mouse germline development; L. Montoliu for discussion of the tyrosinase locus; M. Tuszynski for plasmids and for early support of the project. This work was funded by a Searle Scholar Award from the Kinship Foundation, a Pew Biomedical Scholar Award from the Pew Charitable Trusts, a Packard Fellowship in Science and Engineering from the David and Lucile Packard Foundation, and NIH grant R21GM129448 awarded to K.L.C. E.B. was supported by NIH grant R01GM117321, a Paul G. Allen Frontiers Group Distinguished Investigators Award and a gift from the Tata Trusts in India to TIGS-UCSD and TIGS-India. H.A.G. was supported by a Ruth Stern Graduate Fellowship and by the NIH Cell and Molecular Genetics training grant T32GM724039; V.M.G. was supported by NIH grant DP5OD023098.
Reviewer information
Nature thanks B. Conklin, S. Qi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Affiliations
Contributions
H.A.G., V.M.G., G.P., E.B. and K.L.C. conceived and designed the research; V.M.G. and G.P. designed and cloned the TyrCopyCat transgene and validated mCherry expression in vitro; X.-R.S.X. validated Tyr4a gRNA cleavage activity in vitro; H.A.G. and K.L.C. designed the breeding strategies; H.A.G. acquired and established mouse lines, developed genotyping protocols, and conducted mouse breeding, phenotyping and genotyping; H.A.G. curated the data and analyses; K.L.C. and E.B. supervised the research; K.L.C. wrote the original draft; H.A.G., V.M.G., E.B. and K.L.C. revised subsequent drafts.
Corresponding author
Ethics declarations
Competing interests
V.M.G., E.B. and K.L.C. hold advisory board positions with Synbal. All other authors declare that they have no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Knock-in strategy using the TyrCopyCat-targeting vector.
The U6-Tyr4a gRNA (tyrosinase exon 4 gRNA a) and CMV-mCherry were inserted into the cut site of the Tyr4a gRNA by HDR after CRISPR–Cas9 DSB formation targeted by the Tyr4a gRNA. See Supplementary Methods and Supplementary Figs. 1, 2 for additional details.
Extended Data Fig. 2 Rosa26-cas9 and H11-cas9 constitutive lineages have different numbers of unique NHEJ indels.
Sanger sequencing of the Tyr4a gRNA target exon amplified from tail-tip genomic DNA using TyrHALF2 and TyrHARR2 primers as specified in Supplementary Table 3. Top, a single representative Sanger sequence trace of the bulk PCR product amplified from a Rosa26-cas9;TyrCopyCat-positive F2 mouse (Rosa26 family 1 in Extended Data Table 3) with either major or minor peaks called revealing two distinct alleles. Five Tyrch-positive F3 offspring of this F2 individual each match one of the two alleles (marked 1 (insertion) and 2 (deletion)). Bottom, a single representative sequence trace of the bulk PCR product amplified from an H11-cas9;TyrCopyCat-positive F2 mouse (H11 family 1 in Extended Data Table 3). Alternate alleles cannot be called because of the complexity of overlapping peaks. Five Tyrch-positive F3 offspring each have one of four different alleles (marked 1, 2, 3 and 4). Sequence trace data are representative of all 90 individuals of 5 families of each constitutive strategy described in Extended Data Table 3.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-5 and Supplementary Tables 1-4. Supplementary Methods detail the TyrCopyCat targeted insertion into the mouse genome and genotyping methods for all alleles together with supporting Supplementary Figures. Supplementary Tables include all primers and mouse lines used in this study and information about the distribution of coat colors among F3 parents of each germline strategy.
Rights and permissions
About this article
Cite this article
Grunwald, H.A., Gantz, V.M., Poplawski, G. et al. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 566, 105–109 (2019). https://doi.org/10.1038/s41586-019-0875-2
Received:
Accepted:
Published:
Issue Date:
Further reading
-
Scenario analysis on the use of rodenticides and sex-biasing gene drives for the removal of invasive house mice on islands
Biological Invasions (2020)
-
Spatiotemporal Controllability and Environmental Risk Assessment of Genetically Engineered Gene Drive Organisms from the Perspective of European Union Genetically Modified Organism Regulation
Integrated Environmental Assessment and Management (2020)
-
The ethics of genetic engineering and gene drives in conservation
Conservation Biology (2020)
-
Advances in Molecular Tools and In Vivo Models for the Study of Human Fungal Pathogenesis
Microorganisms (2020)
-
Performance analysis of novel toxin-antidote CRISPR gene drive systems
BMC Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.