Abstract
Plant gene editing is typically performed by delivering reagents such as Cas9 and single guide RNAs to explants in culture. Edited cells are then induced to differentiate into whole plants by exposure to various hormones. The creation of edited plants through tissue culture is often inefficient, time-consuming, works for only limited species and genotypes, and causes unintended changes to the genome and epigenome. Here we report two methods to generate gene-edited dicotyledonous plants through de novo meristem induction. Developmental regulators and gene-editing reagents are delivered to somatic cells of whole plants. This induces meristems that produce shoots with targeted DNA modifications, and gene edits are transmitted to the next generation. The de novo induction of gene-edited meristems sidesteps the need for tissue culture and promises to overcome a bottleneck in plant gene editing.
Access options
Subscription info for Chinese customers
We have a dedicated website for our Chinese customers. Please go to naturechina.com to subscribe to this journal.
Rent or Buy article
Get time limited or full article access on ReadCube.
from$8.99
All prices are NET prices.
Data availability
High-throughput sequencing data have been deposited in the NCBI Sequence Read Archive under the BioProject accession number PRJNA575069. Sanger DNA sequence data are provided as a Supplementary Data Set. Constructs expressing DRs and gene-editing reagents are available from Addgene (plasmids 127210–127230, 133312–133315). Correspondence and requests for materials should be addressed to D.F.V. (voytas@umn.edu).
Change history
07 May 2020
In the supplementary information originally posted for this article, Supplementary Fig. 3d was missing and lettering in Supplementary Fig. 3a was corrupted. The error has been corrected online.
References
-
1.
Barton, M. K. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113 (2010).
-
2.
Gallois, J.-L., Woodward, C., Reddy, G. V. & Sablowski, R. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129, 3207–3217 (2002).
-
3.
Ckurshumova, W., Smirnova, T., Marcos, D., Zayed, Y. & Berleth, T. Irrepressible MONOPTEROS/ARF5 promotes de novo shoot formation. N. Phytol. 204, 556–566 (2014).
-
4.
Smigocki, A. C. & Owens, L. D. Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc. Natl Acad. Sci. USA 85, 5131–5135 (1988).
-
5.
Ebinuma, H., Sugita, K., Matsunaga, E. & Yamakado, M. Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc. Natl Acad. Sci. USA 94, 2117–2121 (1997).
-
6.
Lowe, K. et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015 (2016).
-
7.
Lowe, K. et al. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. Vitr. Cell. Dev. Biol., Plant 54, 240–252 (2018).
-
8.
Nelson-Vasilchik, K., Hague, J., Mookkan, M., Zhang, Z. J. & Kausch, A. Transformation of recalcitrant Sorghum varieties facilitated by Baby Boom and Wuschel2. Curr. Protoc. Plant Biol. 3, e20076 (2018).
-
9.
Altpeter, F. et al. Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016).
-
10.
Bally, J. et al. The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annu. Rev. Phytopathol. 56, 405–436 (2018).
-
11.
Wu, H.-Y. et al. AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10, 19 (2014).
-
12.
Wang, J. & Jiao, Y. Axillary meristem initiation – a way to branch out. Curr. Opin. Plant Biol. 1, 61–66 (2018).
-
13.
Baltes, N. J. et al. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 1, 15145 (2015).
-
14.
Bombarely, A. et al. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 25, 1523–1530 (2012).
-
15.
Qin, G. et al. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17, 471–482 (2007).
-
16.
Jansing, J., Sack, M., Augustine, S. M., Fischer, R. & Bortesi, L. CRISPR/Cas9-mediated knockout of six glycosyltransferase genes in Nicotiana benthamiana for the production of recombinant proteins lacking β-1,2-xylose and core α-1,3-fucose. Plant Biotechnol. J. 17, 350–361 (2019).
-
17.
Phillips, R. L., Kaeppler, S. M. & Olhoft, P. Genetic instability of plant tissue cultures: breakdown of normal controls. Proc. Natl Acad. Sci. USA 91, 5222–5226 (1994).
-
18.
Zhang, D. et al. Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PloS ONE 9, e96879 (2014).
-
19.
Gelvin, S. B. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37 (2003).
-
20.
Hamada, H. et al. An in planta biolistic method for stable wheat transformation. Sci. Rep. 7, 11443 (2017).
-
21.
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
-
22.
Čermák, T. et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29, 1196–1217 (2017).
-
23.
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A. & Voytas, D. F. DNA replicons for plant genome engineering. Plant Cell 26, 151–163 (2014).
-
24.
Doyle, J. J. & Doyle, J. L. Isolation of plant DNA from fresh tissue. Focus 12, 13–15 (1990).
-
25.
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
-
26.
Aronesty, E. Comparison of sequencing utility programs. Open Bioinforma. J. 7, 1–8 (2013).
-
27.
Park, J., Lim, K., Kim, J.-S. & Bae, S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288 (2017).
Acknowledgements
This work was supported by the Hackett Fund of the University of Minnesota; the work on grape was supported by TechAccel. M.F.M. was funded from NIGMS T32-GM008347. We thank M. Leffler for help with the figures and P. Atkins for help with the next-generation sequencing data analysis.
Author information
Affiliations
Contributions
M.F.M. and R.A.N. designed the research, performed experiments, analyzed data and wrote the manuscript. M.V. and M.D.C. assisted with the experiments on potato and grape, respectively. C.G.S. helped to build DNA constructs. D.F.V. supervised the research and wrote the manuscript. All of the authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.F.M., R.A.N. and D.F.V. are named inventors on a patent application pertaining to the technology that was filed by the University of Minnesota. D.F.V. serves as Chief Science Officer for Calyxt, an agricultural biotechnology company that uses gene editing to create new crop varieties. All other authors have no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Generating de novo meristems on seedlings.
a, Wus2- and STM-induced growths on 21–31% of treated seedlings. Few differences in frequency were observed among the different promoters used to express STM, probably because they all drive high expression in N. benthamiana. b–d, Approximately 12 d after initiating Fast-TrACC, callus-like growths formed, varying from one (b) to many (c,d) per seedling. e,f, Over the next 10–14 d, the growths expanded in size and remained in an undifferentiated state or began to differentiate. g,h, Shoot-like structures then formed, which could be excised and moved to rooting medium. The growths depicted are representative of those observed in three independent experiments.
Supplementary Figure 2 Plants with developmental abnormalities.
a–c, Some transgenic plants displayed developmental abnormalities. Shown here are plants derived from treating seedlings with Wus2 and STM. Phenotypes include large numbers of leaves (a), excess branching (b), and misshapen leaves (c). The three plants depicted are representative of plants derived from the 36 growths obtained in the experiment described in Supplementary Fig. 1.
Supplementary Figure 3 Inducing de novo meristems in tomato using Fast-TrACC.
a, To induce de novo meristems in tomato, three combinations of DRs (nos:Wus2 & 35S:STM, nos:Wus2 & CmYLCV:STM, and nos:Wus2 & 35S:ipt) were selected because they effectively induced meristems in N. benthamiana. (Supplementary Table 1). For both combinations of Wus2 & STM, no shoot-like growths formed. b, In contrast, Wus2 & ipt promoted shoot-like growths that ultimately formed fully rooted plants. c, When a luciferase reporter was delivered with the DRs, meristems were induced (black arrow). d, These meristems were positive for luciferase activity (red box and enlarged image shown as inset).
Supplementary Figure 4 Gene edits are transmitted vertically by plants derived from de novo meristems induced by Fast-TrACC.
a, Five gene-edited plants were produced by treating seedlings with Wus2 & ipt, Wus2 & STM or Wus2 alone. The type and amount of editing were determined by Sanger sequencing and TIDE analysis (see Methods). The predicted editing percentage for each PDS gene is shown. b, The types of edits predicted by TIDE are shown as percentages of total editing. Any percentage below 5% was deemed below the limit of detection by TIDE. The first number in the column is indicative of the percentage of that edit in PDS1; the second number is indicative of editing in PDS2. Because of a deletion in 4-2, the sequence could not be properly aligned, and thus no percentages are given for PDS2. See the Supplementary Data Set for DNA sequences of all mutants recovered.
Supplementary Figure 5 Heritability of N. benthamiana PDS editing events created by Fast-TrACC.
a, An example of the seedling phenotypes derived from flowers F4 and F5 of plant 1-7 (see also Supplementary Table 4). The fully green seedlings are healthy; the fully white seedlings do not develop after germination; the chimeric plants are stunted. b, Chimeric seedlings retain the transgene that carries the sgRNA expression cassette. DNA from seedlings was PCR-amplified using primers that specifically amplify the transgene. Chimerism s probably owing to continued mutagenesis at PDS1. The image in panel a is representative of experiments performed once with seeds derived from flowers F4 and F6. The experiment in panel b was performed once.
Supplementary Figure 6 Representative images of phenotypes observed for newly formed tissues at DR delivery sites.
a–d, Many of the phenotypes suggest improper meristem patterning, probably owing to persistent DR expression. b, Some shoots and leaf tissues are white, suggesting biallelic mutations in both PDS homologs. c, Other tissues show a mixture of both green and white phenotypes. d, When floral tissues form on morphologically abnormal tissues, they are abnormal and typically infertile. e, Some shoots appear phenotypically normal. The shoot images are representative of the nine white shoots and 30 distorted shoots obtained in the experiment described in Fig. 5c.
Supplementary Figure 7 Delivery of DRs to grape plants induced phenotypically normal transgenic shoots.
The left image is an example of a grape plant 40 d after inoculation with five A. tumefaciens strains, each carrying a T-DNA vector that expresses a single DR (pMM230-234). Newly formed shoots were generated at inoculation sites. The image on the right shows shoots that were removed from the plant on the left, exposed to luciferin, and imaged for bioluminescence. Bioluminescent tissues indicate newly formed, transgenic shoots that express the luciferase reporter. Images are from a single experiment performed in grape and represent one of six inoculated plants.
Supplementary Figure 8 Delivery of DRs to potato plants induced abnormal shoot phenotypes and transgenic shoots.
a, The potato plant pMKV059-6 displays abnormal shoot formation approximately 100 d after inoculation with an A. tumefaciens strain carrying a T-DNA with ipt and luciferase expression cassettes. b, Luciferase expression is observed in tissues and shoots of plant pMKV059-6. c, Fully transgenic shoots (white arrowheads) are more easily seen after trimming away several wild-type shoots. d, Potato plant pMKV057-1 displays abnormal shooting approximately 100 d after inoculation with an A. tumefaciens strain carrying a T-DNA with ipt, Wus2 and luciferase expression cassettes. e, Luciferase expression is observed in tissues and shoots of plant pMKV057-1. f, Plant pMKV057-1 was trimmed, leaving some transgenic tissue. g, Fully transgenic shoots could be isolated. The images are representative of two experiments performed in potato.
Supplementary Figure 9 Evidence for gene editing in plants derived from the experiment described in Fig. 4b.
Plants were treated with constructs that included a single vector containing Wus2 and STM (Wus2/STM), a single vector containing Wus2 and ipt (Wus2/ipt), or a combination of co-inoculated vectors each containing a single DR (All combination, see Fig. 4 legend). Derived shoots were given individual designators to facilitate sample tracking. Purified genomic DNA was amplified for the PDS2 locus. Amplicons were pooled and submitted for next-generation sequencing. ‘Sequences analyzed’ denotes the observed number of sequences that contained the expected forward and reverse barcodes. ‘Insertions’ denotes the number of sequences observed to have non-specific DNA insertions at the sgRNA target site. ‘Deletions’ denotes the number of sequences observed to have targeted mutations at the sgRNA target site. ‘Indel frequency’ denotes the total number of sequences that were observed to have targeted modifications. ‘Observed mutations’ denotes mutations observed at a frequency >30% of the total. ‘Seed produced’ denotes those sampled shoots that produced seed. With the exception of plant 1-1-5, sequences of the resulting mutations are shown in Fig. 5b. Below the table are the spectra of mutations recovered for plant 1-1-5.
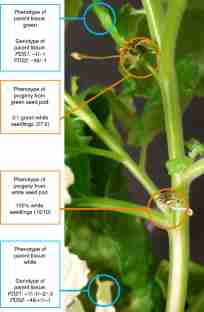
Supplementary Figure 10 Evidence for gene editing in seedlings derived from the white seed pod in Fig. 6.
Observed mutations in the white parental tissues and the progeny of the white seed pod that is shown Fig. 6. Genomic DNA was extracted from parental tissue and seedlings and submitted for Sanger sequencing. Sequences were assessed for mutations by TIDE sequence trace analysis (see Methods). All seedlings were white, as shown in the photo. See the Supplementary Data Set for the DNA sequences of all mutations recovered.
Supplementary Figure 11 Gene-edited seedlings lack integrated T-DNA.
Genomic DNA was extracted from parental white (ParW) and parental green (ParG) tissues of a plant demonstrating targeted editing (Fig. 6), as well as from ten seedlings derived from the white flower (S1-S10; Extended Data Fig. 10). Genomic DNA was also extracted from plants that did not receive the vector (Neg Ctrl 1 and 2) and from leaf tissue infiltrated with the target vector (Pos Ctrl). DNA was amplified using primers specific to the U6 promoter present on the T-DNA (expected 448 bp). New England Biolabs 2-Log DNA Ladder. The experiment was performed once.
Supplementary Figure 12 Evidence for gene editing in seedlings derived from the green seed pod in Fig. 6.
Observed mutations in the green tissues and the progeny from the green seed pod shown in Fig. 6. Genomic DNA was extracted from parental tissue and seedlings and submitted for Sanger sequencing. Sequences were assessed for mutations by TIDE sequence trace analysis (see Methods). The photo shows wild-type and pds seedling phenotypes demonstrating a 3:1 segregation. Note: the 48 bp deletion is an in-frame deletion, which appears to retain PDS function.
Supplementary information
Supplementary Materials
Supplementary Figs. 1–12 and Supplementary Tables 1–4.
Supplementary Data Sets
Sanger DNA sequences
Rights and permissions
About this article
Cite this article
Maher, M.F., Nasti, R.A., Vollbrecht, M. et al. Plant gene editing through de novo induction of meristems. Nat Biotechnol 38, 84–89 (2020). https://doi.org/10.1038/s41587-019-0337-2
Received:
Accepted:
Published:
Issue Date:
Further reading
-
Genetically Modified Plants: Nutritious, Sustainable, yet Underrated
The Journal of Nutrition (2020)
-
Fast-track for engineered plants
Nature Biotechnology (2020)
-
Improving CRISPR‐Cas‐mediated RNA targeting and gene editing using SPLCV replicon‐based expression vectors in Nicotiana benthamiana
Plant Biotechnology Journal (2020)
-
Establishment of the 'imbibed seed piercing' method for Agrobacterium-mediated transformation of jute and flax bast fibre crops via phloem-specific expression of the β-glucuronidase Gene
Industrial Crops and Products (2020)
-
Agrobacterium-mediated delivery of CRISPR/Cas reagents for genome editing in plants enters an era of ternary vector systems
Science China Life Sciences (2020)